The Human Genome and Human Gene Therapy:
The strategy of human gene therapy is the introduction of DNA into human cells to correct inherited genetic deficiencies.
E.g. Severe Combined Immune Deficiency (SCID). One form of SCID results from genetically inherited defects in gene encoding adenosine Deaminase (ADA), an enzyme involved in nucleotide biosynthesis. Another form of SCID arises from defect in cell surface receptor for Interleukin which triggers differentiation. In both case the progenitor stem cells cannot differentiate into mature immune cells like T and B lymphocytes. Children with this disorder are highly susceptible to bacterial and viral infections.
A gene therapy trial initiated in 1999 was successful in correcting a form of SCID caused by defective cytokine receptors. The researchers introduced the corrected gene for γc cytokine receptor subunit in retrovirus and this was again transfected into CD34+ cells (the stem cells that give rise to immune system cells). The transformed cells were placed back into the patient’s bone marrow. The corrected gene conferred a growth over untreated cells. A functioning immune system was detected. Although two of patients developed severe form of leukemia due to retrovirus itself got integrated itself into a chromosome of stem cells resulting in abnormally high expression of hematopoietic oncogene that led to uncontrolled cell growth.
The first human gene therapy trial was carried out at National Institute of Health in Bethesda, Maryland in 1990. The patient was a four year old girl crippled by ADA deficiency. Bone marrow cells from the child were transformed with an engineered retrovirus containing a functional, ADA gene. The transformed cells in vivo were introduced into the patient’s marrow. Four years later, the child was leading a normal life.
Although the gene therapy is absolute care it is associated with various risks. One major impediment proved to be the inefficiency of introducing new genes into cells. Transformation failed in many cells, and the number of transformed cells often proved insufficient to reverse the disorder.
Human gene therapy is not limited to genetic diseases. Cancer cells are being targeted by delivering genes for proteins that might destroy the cell or restore the normal cell division. Immune system cells associated with tumors can be genetically modified to produce tumor necrosis factor (TNF). When these lymphocytes are taken from a cancer patient, modified, and reintroduced, the engineered cells target the tumor, and the TNF they produce causes tumor shrinkage. AIDS may also be treatable with gene therapy; DNA that encodes an RNA molecule complementary to a viral HIV mRNA could be introduced into immune system cells (the targets of HIV). The RNA transcribed from the introduced DNA would pair with the HIV mRNA, preventing its translation and interfering with the virus's life cycle.
Human Genome Project:
Discussion in mid 1980 led to initiation of the Human Genome Project in 1989 but the actual work of sequencing began at 1990. Researchers team first generated a detailed physical map of human genome with clones derived from each chromosome.
 |
| Fig: The Human genome project strategy: Sequenced by shotgun sequencing. |
Human genome project was completed in 2001 and published in April 2003. We have only 25, 000 – 30, 000 genes and 3.9 X 109 bp per haploid genome i.e. 23 chromosomes. 1.1 to 1.4% of our DNA actually encodes proteins. More than 50% of our genome consist of short repeated sequences the vast majority of which about 45% come from transposons. The human genome
can encode 30,000 to 40,000 proteins. There are more than 3 million SNPs. Whole genome contains 10% of Alu elements and present in GC rich region. This project was handled by international collaboration involving groups from the USA, UK, Japan, France, Germany and china known as International Human Genome Sequencing Consortium (IHGSC).
The human and chimpanzee genome differ by only 1.2% at the level of base pairs and even less in gene encoding proteins. This creates 35 million base pair changes.
The overall approach, referred to as hierarchical shotgun sequencing, consisted of fragmenting the entire genome into pieces of approximately 100–200 kb and inserting them into bacterial artificial chromosomes (BACs). The BACs were then positioned on individual chromosomes by looking for marker sequences known as sequence-tagged sites STSs), whose locations had been already determined. STSs are short (usually < 500 bp), unique genomic loci for which a PCR assay is available. Clones of the BACs were then broken into small fragments (shotgunning). Each fragment was then sequenced, and computer algorithms were used that recognized matching sequence information from overlapping fragments to piece together the complete sequence.
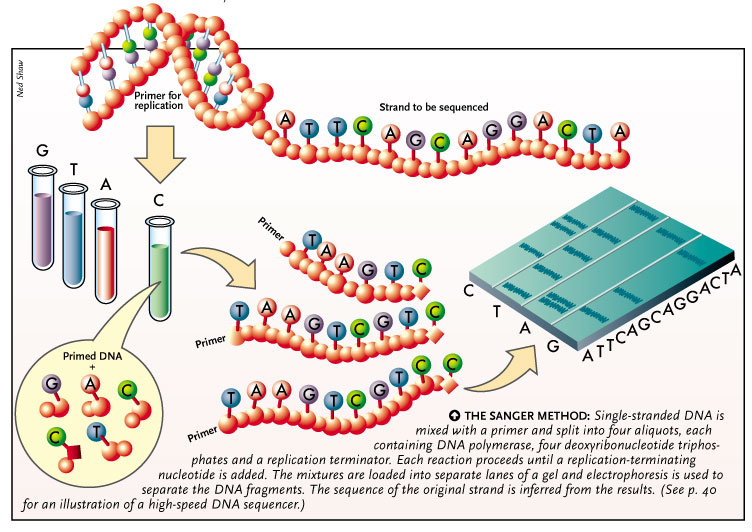
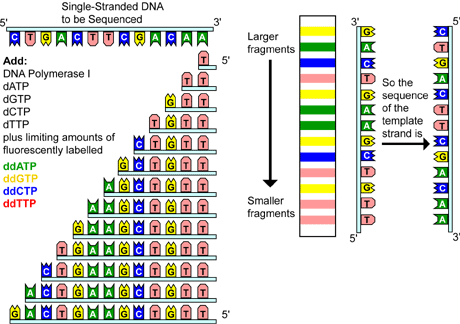
On the basis of STS, PCR based sequencing was done using primers that recognize the vector terminal sequence and using labeled dNTPs. ddNTP was used to terminate the chain elongation process to generate short fragments formed due to addition of ddNTP in the complementary site. These fragments were then sequences and assembled to determine the overlapping bases on the basis of these overlap fragments from entire genome fragments were joined to produce final sequence.
For More information on Human Genome Project, Visit:
(Source: Lehninger's Textbook of Biochemistry)