Visualization of specific DNA, RNA or Protein out of contaminants can be done by blotting techniques like those shown in figure above. These procedure help to determine the number of copies of genes present in the given tissue or whether there are any gross alteration in a gene (deletions, insertions, or rearrangements) because any alteration will change the size which can be resolved by electrophoresis.
 |
| Fig. Blotting Technique |
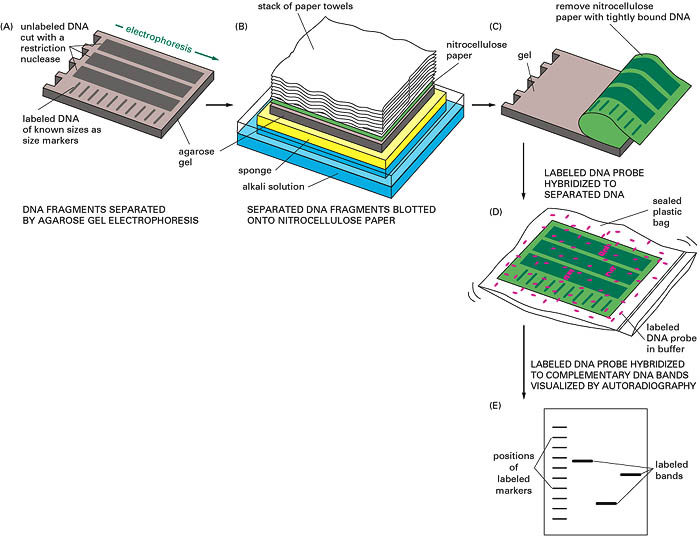
This is a DNA blot transfer. It combines the use of restriction enzyme, electrophoresis and DNA probes. DNA isolated from cell line or tissue is digested with one or more restriction enzymes. This mixture is pipetted into a well in an agarose or polyacrylamide gel and electrophoresed. DNA being negatively charged migrates towards the anode; the smaller fragments move rapidly. (The length of fragments can be determined by comparison of the position of band relative to standard fragment of known size). After this, the DNA is denatured by exposure to mild alkali and transferred to nitrocellulose or nylon paper, resulting in an exact replica of the pattern on the gel. The DNA is bound to the paper by exposure to heat or UV, and the paper is then exposed to the labelled cDNA probe, which hybridizes the complementary fragments on the filter. After through washing the paper is exposed to x-ray film, which produces specific bands corresponding to the DNA fragment that recognized the sequences in the cDNA probe.
 |
| Fig. Western blotting |
Northern Blot:
This is similar to southern blot. RNA is subjected to electrophoresis before blot transfer.
Western Blot:
This is for proteins. Proteins are electrophoresed and transferred to special paper that avidly binds macromolecules and then probed with specific antibody or other probe molecules.
 |
| Fig. Overall Blotting Technique |
It is one of the methods used to identify a particular DNA fragment from a plasmid gene library. Large clones are grown on agar plate as colonies. These colonies are overlaid with nitrocellulose filter paper where cells will stick. These cells are fixed by heat or UV, and subsequent NaOH treatment will lyse the cell and denatures DNA so that it is available to hybridize with probe. A radioactive probe is added to filter and the hybrid complex is localized by exposing the filter to x-ray film or imaging screen. By matching the spot on the autoradiograph to a colony, the colonies can be picked from the plate. Similar process is used to identify fragments in phase library.
PCR secreening of gene libraries
Hybrid select technique:
This is for screening of cDNA libraries. Here plasmid colonies harboring cDNA is plated on medium. DNA is extracted from each colony denatured and immobilized in membrane. It is exposed to total cellular mRNA and allowed to hybridize with complementary strand. Bound mRNA is eluted from each membrane and is directed to in vitro translation. The proteins produced are then characterized and the clone containing its corresponding cDNA is isolated. They are then further grown for further analysis.
Screening expression cDNA libraries:
This is also for screening of cDNA libraries. Here the cDNA is inserted along with gene to produce fusion protein which can be assayed. This recombinant vectors are plated and allowed to express the protein of interest. The colonies are transferred to membrane and incubated with antibody specific for that protein which will produce color. This allows visualization of plaque or colony that contains the cloned cDNA for that protein which may then be picked from the agar plate and pure preparations grown for analysis.
(Source: Lippincott's Illustrated Biochemistry, Practical Biochemistry and Harper's Biochemistry)


No comments:
Post a Comment